Home » Health News »
Network-based approaches open a new avenue to classify and treat rare diseases
Network-based approaches often unveil what remains concealed—this holds true in medical research as well. For several years, CeMM Adjunct Principal Investigators Kaan Boztug, Director of St. Anna Children’s Cancer Research Institute, and Jörg Menche, a professor at the University of Vienna and Max Perutz Labs have been working to gain a better systemic and molecular understanding of rare diseases, congenital immune disorders, and congenital inflammatory disorders by using network-based methods.
In their latest study led by the study’s first author Julia Guthrie, the researchers successfully identified new molecular and mechanistic similarities between rare immune system disorders by examining the high degree of interconnectedness of molecular interactions, leading to their reclassification. By comparing their results to clinical data, the researchers demonstrated that patients with diseases within a classification group also responded to the same medications.
New classification enables more targeted therapies
For their study, the researchers examined around 200 rare immune disorders with inflammatory phenotypes. The network-based analysis of protein-protein interactions revealed similarities in the molecular mechanisms behind these diseases. Through these analyses, the diseases were reclassified, and the researchers subsequently calculated which therapies could yield the best results for each respective group.
“Compared to existing clinical data, the new disease classification allows for a much better prediction of promising therapies compared to the previous approach. Network biology allows us to gain deeper insights into the intricate interplay between the immune system and diseases. This, in turn, enables us to develop more targeted and personalized approaches for diagnosis and treatment,” explained co-study leader Kaan Boztug.
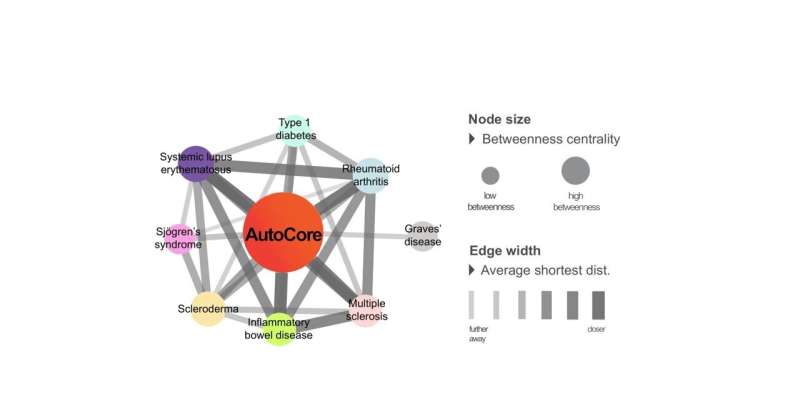
Similar patterns in autoimmune and autoinflammatory diseases
The results also indicate that numerous autoimmune and autoinflammatory diseases such as chronic inflammatory disorders, multiple sclerosis, systemic lupus erythematosus, and type 1 diabetes are closely linked.
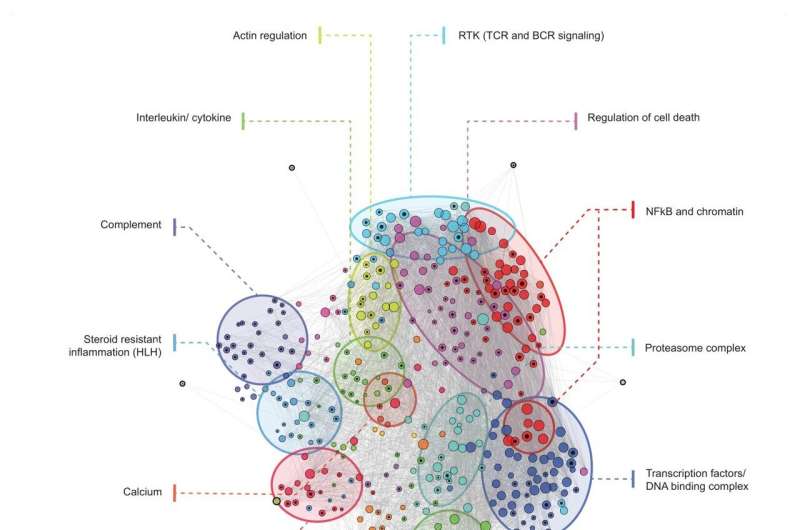
The study’s first author Julia Guthrie explained, “We were able to identify a group of key genes and their interaction partners that are central to homeostasis. We refer to this network of key genes as ‘AutoCore’. In autoimmune and autoinflammatory diseases, the AutoCore resides right at the center of the associated genes. Additionally, we identified 19 other subgroups that are intended to provide us with better insights into homeostasis and immune system deregulation.”
Taking a broader perspective
While conventional approaches often categorize immune system disorders according to specific body regions and thus view them in isolation, a systemic approach aims to offer a more detailed picture of underlying mechanisms.
Co-study leader Jörg Menche explained, “We increasingly recognized the conceptual and practical limitations of the traditional paradigm of ‘one gene, one disease’ in the research of rare diseases. This hinders our understanding of the complex molecular network through which the components of the immune system are orchestrated.”
“Therefore, we developed a visualization in the form of a multidimensional network that depicts all currently known monogenic immune defects underlying autoimmunity and autoinflammation, as well as their molecular interactions. As a result, we can see how closely genes are interconnected in rare diseases.”
The acquired data also provide a crucial foundation for identifying better treatment options for specific groups of disorders.
The findings are published in the journal Science Advances.
More information:
Julia Guthrie et al, AutoCore: a network-based definition of the core module of human autoimmunity and autoinflammation, Science Advances (2023). DOI: 10.1126/sciadv.adg6375. www.science.org/doi/10.1126/sciadv.adg6375
Journal information:
Science Advances
Source: Read Full Article



